Interesting recent developments:
Celts and hg. R1b
Gauls
Recent paper (behind paywall) Multi-scale archaeogenetic study of two French Iron Age communities: From internal social- to broad-scale population dynamics, by Fischer et al. J Archaeol Sci (2019).
In it, Fischer and colleagues update their previous data for the Y-DNA of Gauls from the Urville-Nacqueville necropolis, Normandy (ca. 300-100 BC), with 8 samples of hg. R, at least 5 of them R1b. They also report new data from the Gallic cemetery at Gurgy ‘Les Noisats’, Southern Paris Basin (ca. 120-80 BC), with 19 samples of hg. R, at least 13 of them R1b.
In both cases, it is likely that both communities belonged (each) to the same paternal lineages, hence the patrilocal residence rules and patrilineality described for Gallic groups, also supported by the different maternal gene pools.
The interesting data would be whether these individuals were of hg. R1b-L21, hence mainly local lineages later replaced or displaced to the west, or – a priori much more likely – of some R1b-U152 and/or R1b-DF27 subclades from Central Europe that became less and less prevalent as Celts expanded into more isolated regions south of the Pyrenees and into the British Isles. Such information is lacking in the paper, probably due to the poor coverage of the samples.
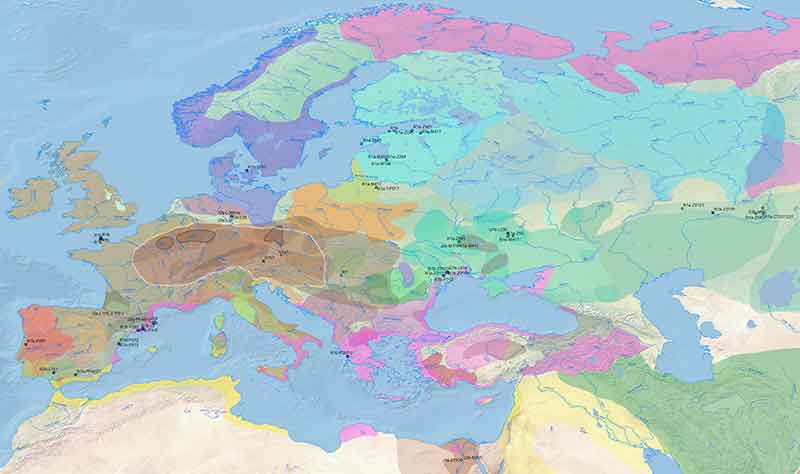
Other Celts
As for early Celts, we already have:
Celtiberians from the Basque Country (one of hg. I2a) and likely Celtic genetic influence in north-east Iberia (all R1b), where Iberian languages spread later, showing that Celts expanded from some place in Central Europe, probably already with the Urnfield culture (ca. 1300 BC on).
Two Hallstatt samples from Bylany, Bohemia (ca. 836-780 BC), by Damgaard et al. Nature (2018), one of them of hg. R1b-U152.

Another Hallstatt HaC/D1 sample from Mittelkirchen, Austria (ca. 850-650/600), by Kiesslich et al. (2012), with predicted hg. G2a (see Athey’s haplogroup prediction).
One sample of early La Tène culture A from Putzenfeld am Dürrnberg, Hallein, Austria (ca 450–380 BC), by Kiesslich et al. (2012), with predicted hg. R1b (see Athey’s haplogroup prediction).
NOTE. For potential unreliability of haplogroup prediction with Whit Atheys’ haplogroup predictor, see e.g. Zhang et al. (2017).

Three Britons from Hinxton, South Cambridgeshire (ca. 170 BC – AD 80) from Schiffels et al. (2016), two of them of local hg. R1b-S461.
Indirectly, data of Vikings by Margaryan et al. (2019) from the British Isles and beyond show hg. R1b associated with modern British-like ancestry, also linked to early “Picts”, hence likely associated with Britons even after the Anglo-Saxon settlement. Supporting both (1) my recent prediction of hg. R1b-M167 expanding with Celts and (2) the reason for its presence among modern Scandinavians, is the finding of the first ancient sample of this subclade (VK166) among the Vikings of St John’s College Oxford, associated with the ‘St Brice’s Day Massacre’ (see Margaryan et al. 2019 supplementary materials).
The R1b-M167 sample shows 23.5% British-like ancestry, hence autosomally closer to other local samples (and related to the likely Picts from Orkney) than to some of his deceased partners at the site. Other samples with sizeable British-like ancestry include VK177 (32.6%, hg. R1b-U152), VK173 (33.3%, hg. I2a1b1a), or VK150 (25.6%, hg. I2a1b1a), while typical Germanic subclades like I1 or R1b-U106 – which may be associated with Anglo-Saxons, too – tend to show less.
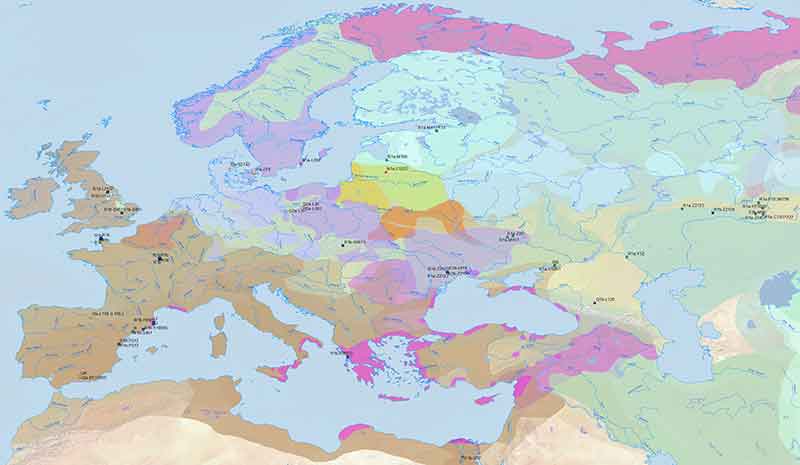
I remember some commenter asking recently what would happen to the theory of Proto-Indo-European-speaking R1b-rich Yamnaya culture if Celts expanded with hg. R1a, because there were only one hg. R1b and one (possibly) G2a from Hallstatt. As it turns out, they were mostly R1b. However, the increasingly frequent obsession of searching for specific haplogroups and ancestry during the Iron Age and the Middle Ages is weird, even as a desperate attempt, because:
- it is evident that the more recent the ancient DNA samples are, the more they are going to resemble modern populations of the same area, so ancient DNA would become essentially useless;
- cultures from the early Iron Age onward (and even earlier) were based on increasingly complex sociopolitical systems everywhere, which is reflected in haplogroup and ancestry variability, e.g. among Balts, East Germanic peoples, Slavs (of hg. E1b-V13, I2a-L621), or Tocharians.
In fact, even the finding of hg. R1b among Celts of central and western Europe during the Iron Age is rather unenlightening, because more specific subclades and information on ancestry changes are needed to reach any meaningful conclusion as to migration vs. acculturation waves of expanding Celtic languages, which spread into areas that were mostly Indo-European-speaking since the Bell Beaker expansion.
Afanasevo ancestry in Asia
Wang and colleagues continue to publish interesting analyses, now in the preprint Inland-coastal bifurcation of southern East Asians revealed by Hmong-Mien genomic history, by Xia et al. bioRxiv (2019).
Interesting excerpt (emphasis mine):
Although the Devil’s Cave ancestry is generally the predominant East Asian lineage in North Asia and adjacent areas, there is an intriguing discrepancy between the eastern [Korean, Japanese, Tungusic (except northernmost Oroqen), and Mongolic (except westernmost Kalmyk) speakers] and the western part [West Xiōngnú (~2,150 BP), Tiānshān Hun (~1,500 BP), Turkic-speaking Karakhanid (~1,000 BP) and Tuva, and Kalmyk]. Whereas the East Asian ancestry of populations in the western part has entirely belonged to the Devil’s Cave lineage till now, populations in the eastern part have received the genomic influence from an Amis-related lineage (17.4–52.1%) posterior to the presence of the Devil’s Cave population roughly in the same region (~7,600 BP)12. Analogically, archaeological record has documented the transmission of wet-rice cultivation from coastal China (Shāndōng and/or Liáoníng Peninsula) to Northeast Asia, notably the Korean Peninsula (Mumun pottery period, since ~3,500 BP) and the Japanese archipelago (Yayoi period, since ~2,900 BP)2. Especially for Japanese, the Austronesian-related linguistic influence in Japanese may indicate a potential contact between the Proto-Japonic speakers and population(s) affiliating to the coastal lineage. Thus, our results imply that a southern-East-Asian-related lineage could be arguably associated with the dispersal of wet-rice agriculture in Northeast Asia at least to some extent.

In this case, the study doesn’t compare Steppe_MLBA, though, so the findings of Afanasievo ancestry have to be taken with a pinch of salt. They are, however, compared to Namazga, so “Steppe ancestry” is there. Taking into account the limited amount of Yamnaya-like ancestry that could have reached the Tian Shan area with the Srubna-Andronovo horizon in the Iron Age (see here), and the amount of Yamnaya-like ancestry that appears in some of these populations, it seems unlikely that this amount of “Steppe ancestry” would emerge as based only on Steppe_MLBA, hence the most likely contacts of Turkic peoples with populations of both Afanasievo (first) and Corded Ware-derived ancestry (later) to the west of Lake Baikal.
(1) The simplification of ancestral components into A vs. B vs. C… (when many were already mixed), and (2) the simplistic selection of one OR the other in the preferred models (such as those published for Yamnaya or Corded Ware), both common strategies in population genomics pose evident problems when assessing the actual gene flow from some populations into others.
Also, it seems that when the “Steppe”-like contribution is small, both Yamnaya and Corded Ware ancestry will be good fits in admixed populations of Central Asia, due to the presence of peoples of EHG-like (viz. West Siberia HG) and/or CHG-like (viz. Namazga) ancestry in the area. Unless and until these problems are addressed, there is little that can be confidently said about the history of Yamnaya vs. Corded Ware admixture among Asian peoples.
Maps, maps, and more maps
As you have probably noticed if you follow this blog regularly, I have been experimenting with GIS software in the past month or so, trying to map haplogroups and ancestry components (see examples for Vikings, Corded Ware, and Yamnaya). My idea was to show the (pre)historical evolution of ancestry and haplogroups coupled with the atlas of prehistoric migrations, but I have to understand first what I can do with GIS statistical tools.
My latest exercise has been to map modern haplogroup distribution (now added to the main menu above) using data from the latest available reports. While there have been no great surprises – beyond the sometimes awful display of data by some papers – I think it is becoming clearer with each new publication how wrong it was for geneticists to target initially those populations considered “isolated” – hence subject to strong founder effects – to extrapolate language relationships. For example:
- The mapping of R1b-M269, in particular basal subclades, corresponds nicely with the Indo-European expansions.
- There is no clear relationship of R1b, not even R1b-DF27 (especially basal subclades), with Basques. There is no apparent relationship between the distribution of R1b-M269 and some mythical non-Indo-European “Old Europeans”, like Etruscans or Caucasian speakers, either.
- Basal R1a-M417 shows an interesting distribution, as do maps of basal Z282 and Z93 subclades, despite the evident late bottlenecks and acculturation among Slavs.
- The distribution of hg. N1a-VL29 (and other N1a-L392 subclades) is clearly dissociated from Uralic peoples, and their expansion in the whole Baltic Sea during the Iron Age doesn’t seem to be related to any specific linguistic expansion.
- Even the most recent association in Post et al. (2019) with hg. N1a-Z1639 – due to the lack of relationship of Uralic with N1a-VL29 – seems like a stretch, seeing how it probably expanded from the Kola Peninsula and the East Urals, and neither the Lovozero Ware nor forest hunter-fishers of the Cis- and Trans-Urals regions were Uralic-speaking cultures.
- The current prevalence of hg. R1b-M73 supports its likely expansion with Turkic-speaking peoples.
- The distribution of haplogroup R1b-V88 in Africa doesn’t look like it was a mere founder effect in Chadic peoples – although they certainly underwent a bottleneck under it.
- The distribution of R1a-M420 (xM198) and hg. R1b-M343 (possibly not fully depicted in the east) seem to be related to expansions close to the Caucasus, supporting once more their location in Eastern Europe / West Siberia during the Mesolithic.
- The mapping of E1b-V13 and I-M170 (I haven’t yet divided it into subclades) are particularly relevant for the recent eastward expansion of early Slavic peoples.

All in all, modern haplogroup distribution might have been used to ascertain prehistoric language movements even in the 2000s. It was the obsession with (and the wrong assumptions about) the “purity” of certain populations – say, Basques or Finns – what caused many of the interpretation problems and circular reasoning we are still seeing today.
I have also updated maps of Y-chromosome haplogroups reported for ancient samples in Europe and/or West Eurasia for the Early Eneolithic, Early Chalcolithic, Late Chalcolithic, Early Bronze Age, Middle Bronze Age, Late Bronze Age, Early Iron Age, Late Iron Age, Antiquity, and Middle Ages.
Haplogroup inference
I have also tried Yleaf v.2 – which seems like an improvement over the infamous v.1 – to test some samples that hobbyists and/or geneticists have reported differently in the past. I have posted the results in this ancient DNA haplogroup page. It doesn’t mean that the inferences I obtain are the correct ones, but now you have yet another source to compare.
Not many surprises here, either:
- M15-1 and M012, two Proto-Tocharians from Shirenzigou, are of hg. R1b-PH155, not R1b-M269.
- I0124, the Samara HG, is of hg. R1b-P297, but uncertain for both R1b-M73 and R1b-M269.
- I0122, the Khvalynsk chieftain, is of hg. R1b-V1636.
- I2181, the Smyadovo outlier of poor coverage, is possibly of hg. R, and could be of hg. R1b-M269, but could also be even non-P.
- I6561 from Alexandria is probably of hg. R1a-M417, likely R1a-Z645, maybe R1a-Z93, but can’t be known beyond that, which is more in line with the TMRCA of R1a subclades and the radiocarbon date of the sample.
- I2181, the Yamnaya individual (supposedly Pre-R1b-L51) at Lopatino II is R1b-M269, negative for R1b-L51. Nothing beyond that.
You can ask me to try mapping more data or to test the haplogroup of more samples, provided you give me a proper link to the relevant data, they are interesting for the subject of this blog…and I have the time to do it.
Related
- European hydrotoponymy (VI): the British Isles and non-Indo-Europeans
- European hydrotoponymy (I): Old European substrate and its relative chronology
- European hydrotoponymy (II): European hydrotoponymy (II): Basques and Iberians after Lusitanians and “Ligurians”
- Haplogroup R1b-M167/SRY2627 linked to Celts expanding with the Urnfield culture
- Aquitanians and Iberians of haplogroup R1b are exactly like Indo-Iranians and Balto-Slavs of haplogroup R1a
- Yamnaya ancestry: mapping the Proto-Indo-European expansions
- Volga Basin R1b-rich Proto-Indo-Europeans of (Pre-)Yamnaya ancestry
- Corded Ware ancestry in North Eurasia and the Uralic expansion
- The genetic and cultural barrier of the Pontic-Caspian steppe – forest-steppe ecotone
- On the origin of haplogroup R1b-L51 in late Repin / early Yamna settlers