Whole mitochondrial genome diversity in two Hungarian populations, Malyarchuk et al. Mol Genet Genomics (2018).
Abstract:
Complete mitochondrial genomics is an effective tool for studying the demographic history of human populations, but there is still a deficit of mitogenomic data in European populations. In this paper, we present results of study of variability of 80 complete mitochondrial genomes in two Hungarian populations from eastern part of Hungary (Szeged and Debrecen areas). The genetic diversity of Hungarian mitogenomes is remarkably high, reaching 99.9% in a combined sample. According to the analysis of molecular variance (AMOVA), European populations showed a low, but statistically significant level of between-population differentiation (Fst = 0.61%, p = 0), and two Hungarian populations demonstrate lack of between-population differences. Phylogeographic analysis allowed us to identify 71 different mtDNA sub-clades in Hungarians, sixteen of which are novel. Analysis of ancestry-informative mtDNA sub-clades revealed a complex genetic structure associated with the genetic impact of populations from different parts of Eurasia, though the contribution from European populations is the most pronounced. At least 8% of ancestry-informative haplotypes found in Hungarians demonstrate similarity with East and West Slavic populations (sub-clades H1c23a, H2a1c1, J2b1a6, T2b25a1, U4a2e, K1c1j, and I1a1c), while the influence of Siberian populations is not so noticeable (sub-clades A12a, C4a1a, and probably U4b1a4).
Interesting excerpt:
Our analysis of ancestry-informative mtDNA sub-clades revealed a complex genetic structure associated with the genetic impact of populations from different parts of Europe. At least 8% of ancestry-informative haplotypes found in Hungarians demonstrate similarity with East (Russians and Ukrainians) and West (Poles and Slovaks) Slavic populations (sub-clades H1c23a, H2a1c1, J2b1a6, T2b25a1, U4a2e, K1c1j, and I1a1c). This observation is consistent with the results of mtDNA studies of medieval populations living in the Hungarian-Slavic contact zone of the Carpathian Basin in the 9th–12th centuries AD (Csákyová et al. 2016). Taken together, these data confirm earlier historical and archaeological reports on mixed populations of medieval Slavs and Magyars, based on the research into cemeteries discovered in Central Europe (Csősz et al. 2016; Csákyová et al. 2016). On the other hand, we cannot confirm the Hungarian-Slavic contacts using molecular dating of the identified mtDNA sub-clades, since their age exceeds the estimated time of the contact period and varies from 1.3 kya (for K1c1j) to 5.2 kya (for T2b25a1) (Figure S1). One of an issue may be sample size problem, because some haplotypes may be missed in the sampling, and this can lead to an overestimate of the age of the mtDNA sub-clade (Richards et al. 2000).
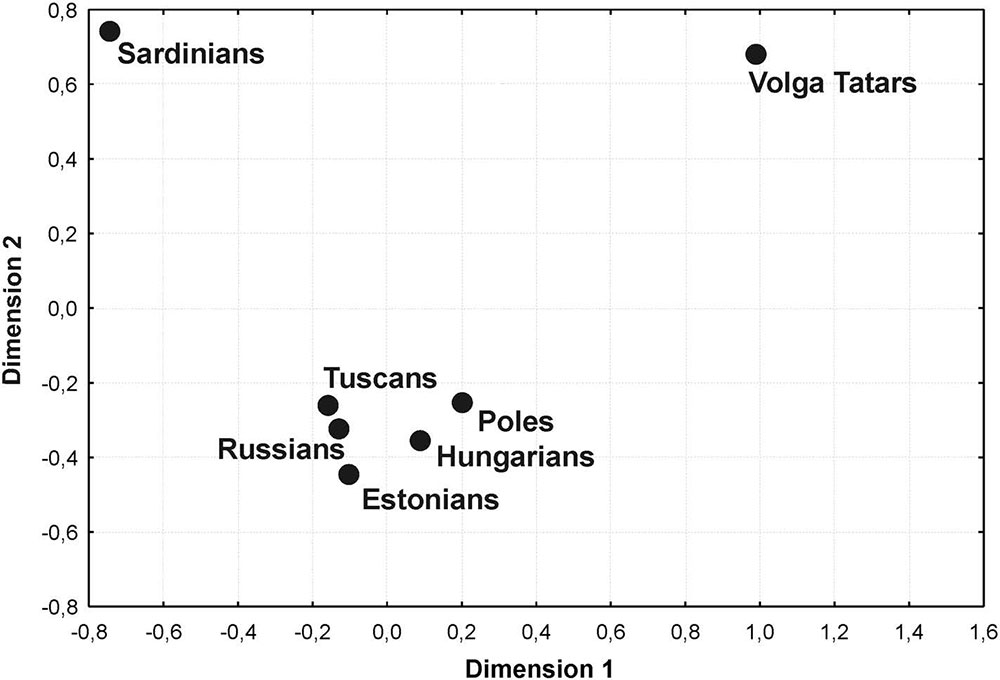
However, it is known that the evolutionary ages of most mtDNA lineages specific to Eastern and Central Europeans correspond to approximately 4 kya (from 2.3 to 5.9 kya) (Malyarchuk et al. 2008, 2017; Mielnik-Sikorska et al. 2013; Översti et al. 2017), thus coinciding with the time of the Bronze Age expansion of Eastern Europeans in accordance with the Kurgan model established by archaeologists and paleogeneticists (Gimbutas 1971; Allentoft et al. 2015; Haak et al. 2015). Thus, similar haplotypes among Hungarians and Slavs and other European ethnic groups can be a reflection of the common genetic substratum which predates the formation of the most modern European populations. Therefore, mtDNA sub-clades H5a1m, T2a1c, and W3a1d1 (with the ages varying from 2.6 to 3.9 kya, based on complete mtDNA mutation rate), which are shared by Hungarians and Finno-Ugric peoples, such as Estonians and Finns, may testify these pan-European relationships (Figure S1). Another example is the sub-clade J2b1a6, which unites the mtDNA haplotypes of the ancient and modern population of Eastern and Central Europe from the Iron Age to the present (Figure S1).
Related:
- First Hungarian ruling dynasty, the Árpáds, of Y-DNA haplogroup R1a
- Admixture of Srubna and Huns in Hungarian conquerors
- Modern Hungarian mtDNA more similar to ancient Europeans than to Hungarian conquerors
- Oldest N1c1a1a-L392 samples and Siberian ancestry in Bronze Age Fennoscandia
- Consequences of Damgaard et al. 2018 (III): Proto-Finno-Ugric & Proto-Indo-Iranian in the North Caspian region
- The concept of “Outlier” in Human Ancestry (III): Late Neolithic samples from the Baltic region and origins of the Corded Ware culture
- Genetic prehistory of the Baltic Sea region and Y-DNA: Corded Ware and R1a-Z645, Bronze Age and N1c
- More evidence on the recent arrival of haplogroup N and gradual replacement of R1a lineages in North-Eastern Europe
- Another hint at the role of Corded Ware peoples in spreading Uralic languages into north-eastern Europe, found in mtDNA analysis of the Finnish population
- New Ukraine Eneolithic sample from late Sredni Stog, near homeland of the Corded Ware culture