Review (behind paywall) The genetic makings of South Asia, by Metspalu, Monda, and Chaubey, Current Opinion in Genetics & Development (2018) 53:128-133.
Interesting excerpts (emphasis mine):
(…) the spread of agriculture in Europe was a result of the demic diffusion of early Anatolian farmers, it was discovered that the spread of agriculture to South Asia was mediated by a genetically completely different farmer population in the Zagros mountains in contemporary Iran (IF). The ANI-ASI cline itself was interpreted as a mixture of three components genetically related to Iranian agriculturalists, Onge and Early and Middle Bronze Age Steppe populations (Steppe_EMBA).
The first ever autosomal aDNA from South Asia comes from Northern Pakistan (Swat Valley, early Iron Age). This study presented altogether 362 aDNA samples from the broad South and Central Asia and contributes substantially to our understanding of the evolutionary past of South and Central Asia. The study redefines the three genetic strata that form the basis of the Indian Cline. The Indus Periphery (IP) component is composed of (varying proportions of): first, IF, second, Ancient Ancestral South Asians (AASI), which represents an ancient branch of human genetic variation in Asia arising from a population split contemporaneous with the splits of East Asian, Onge and Australian Aboriginal ancestors and third, West_Siberian Hunter gatherers (WS_HG).
The authors argue that IP could have formed the genetic base of the Indus Valley Civilization (IVC). Upon the collapse of the IVC IP contributes to the formation of both ASI and ANI. ASI is formed as IP admixes further with AASI. ANI in turn forms when IP admixes with the incoming Middle and Late Bronze Age Steppe (Steppe_MLBA) component, (rather than the Steppe_EMBA groups suggested earlier)
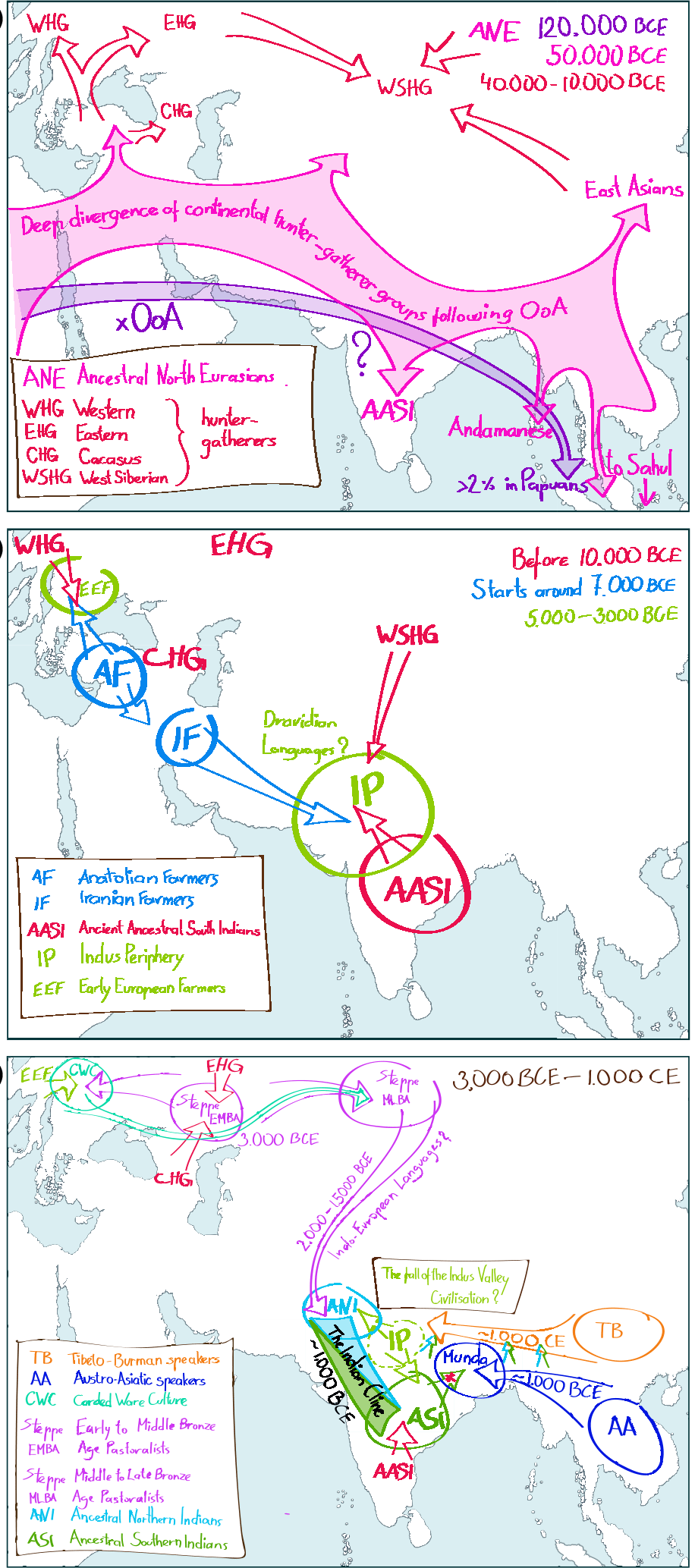
Dating of the arrival of the Austro-Asiatic speakers in South Asia-based on Y chromosome haplogroup O2a1-M95 expansion estimates yielded dates between 3000 and 2000 BCE [30]. However, admixture LD decay-based approach on genome-wide data suggests the admixture between South Asian and incoming Austro-Asiatic speakers occurred slightly later between 1800 and 0 BCE (Tätte et al. submitted). It is interesting that while the mtDNA variants of the Mundas are completely South Asian, the Y chromosome variation is dominated at >60% by haplogroup O2a which is phylogeographically nested in East Asian-specific paternal lineages.
In India, the speakers of Tibeto-Burman (TB) languages live in the Seven Sisters States in Northeast India and in the very north of the country. Genetically they show a clear East Asian origin and around 20% of subsequent admixture with South Asians within the last 1000 years.The genetic flavour of East Asia in TB is different from that in Munda speakers as the best surrogates for the East Asian admixing component are contemporary Han Chinese.
I found the simplistic migration maps especially interesting to illustrate ancient population movements. The emergence of EHG is supposed to involve a WHG:ANE cline, though, and this isn’t clear from the map. Also, there is new information on what may be at the origin of WHG and Anatolian hunter-gatherers.
From the recent Reich’s session on South Asia at ISBA 8:

– Some more certainty about primary window of steppe ancestry injection into S. Asia: 2000-1500 BC
Alexander M. Kim
Very refreshing this morning to hear a senior academic say “we were wrong” – David Reich @Harvard on the genomics of South + Central Asia (with some highly problematic terminology!) #ISBA8 @MPI_SHH #genomics #archsci #indus #archaeogenetics pic.twitter.com/26Zez21VNE
— Sam Leggett (@samleggs22) September 19, 2018
Featured image: map of South Asian languages from http://llmap.org.
Related
- Palaeolithic Caucasus samples reveal the most important component of West Eurasians
- Expansion of haplogroup G2a in Anatolia possibly associated with the Mature Aceramic period
- Steppe and Caucasus Eneolithic: the new keystones of the EHG-CHG-ANE ancestry in steppe groups
- Munda admixture happened probably during the ANI-ASI mixture
- No large-scale steppe migration into Anatolia; early Yamna migrations and MLBA brought LPIE dialects in Asia
- Early Indo-Iranian formed mainly by R1b-Z2103 and R1a-Z93, Corded Ware out of Late PIE-speaking migrations
- South-East Asia samples include shared ancestry with Jōmon
- Model for the spread of Transeurasian (Macro-Altaic) communities with farming
- Genomics reveals four prehistoric migration waves into South-East Asia